This is the official YouTube channel of the Ohue Laboratory. It features research introductions and videos of presentations at international conferences.

Research Highlights
- Development of the protein-protein interaction (PPI) prediction system MEGADOCK
- Development of MEGADOCK-Web, a comprehensive PPI prediction database
- Development of PPI prediction technologies for membrane proteins
- Applications of ultrafast docking calculations in structural biology
- Utilization of AlphaFold2 for protein structure prediction
- Prediction of drug efficacy, activity, and ADMET for small molecules
- Development of statistical models to represent “drug-likeness”
- Development of statistical models to represent “PPI-inhibitor-likeness”
- Development of LBDD screening methods using learning to rank
- Prediction of drug-target interactions using deep learning
- Research on applicability domains for drug-target interaction prediction
- Technology development for peptide screening for drug discovery
- Protein-peptide complex analysis and sequence design using AlphaFold2
- Prediction of protein-cyclic peptide complexes using AlphaFold2
- Structure-based virtual screening using AlphaFold2/3
- GPU supercomputing development using TSUBAME 4.0 and ABCI
- High-parallel computation using Fugaku
- Estimation of high-affinity molecules by binding free energy calculations
- Research on parallel computation techniques for binding free energy calculations
- Efficiency improvement of binding free energy calculations using active learning
- Research on nucleic acid sequence design for drug discovery
- Bioactivity prediction of natural products and derivatives
- Development of antibody design support technologies
- Development of PROTAC design support technologies
- Development of Molecular Glue design support technologies
The above are just examples. Ohue Lab conducts a wide range of research in bioinformatics and drug discovery.
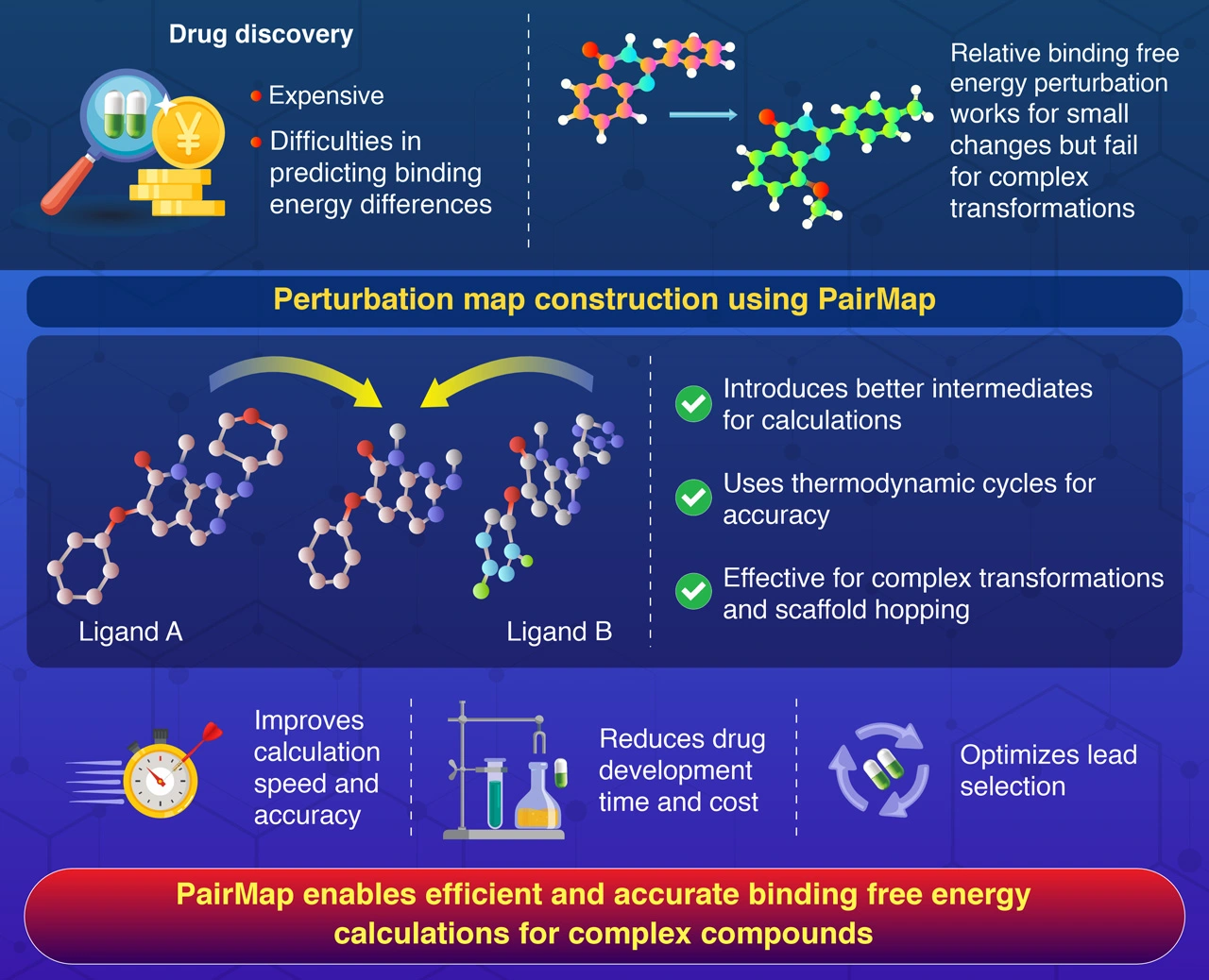
Free Energy Perturbations for Drug Discovery
Drug discovery is a costly and time-intensive process, with binding free energy calculations between the potential drug molecule and the target being crucial for reducing drug discovery costs, streamlining the process and avoiding dead-end leads. Researchers are left with the challenge of predicting how strongly a potential drug molecule will bind to its target, so there is a high demand for computational approaches that can design compounds that bind effectively to the target molecules. However, traditional methods to calculate free energy binding struggle when faced with large chemical transformations or significant molecular rearrangements.
This scenario has led to the development of PairMap, a groundbreaking computational approach developed to address these challenges head-on. Relative binding free energy perturbation (RBFEP) calculations have long been used to predict the binding affinities of chemically similar compounds. While effective for small structural changes, these calculations falter when tasked with large transformations, leading to high errors and computational inefficiencies. PairMap overcomes this limitation by systematically introducing intermediate compounds to create a step-by-step pathway between the two molecules in question. This novel approach minimizes calculation errors, improves convergence, and reduces computational costs.
This study, a joint effort between Ohue Lab and Alivexis, Inc., is a highlight of their close partnership aimed at advancing cutting-edge research. Published online in the Journal of Chemical Information and Modeling on January 12, 2025, the study showcases groundbreaking developments in drug discovery. “PairMap has the potential to redefine drug discovery. By incorporating well-constructed intermediates and thermodynamic cycles, we’ve achieved an unparalleled level of precision in our energy predictions,” says Ohue.
Related Publications:
- Furui K, et al., J. Chem. Inf. Model., 2025: Journal website | PubMed | GitHub
- Furui K, Ohue M. J. Supercomputing, 2024. Journal website | GitHub
Development and Application of MEGADOCK
We are developing software that predicts protein-protein interactions (PPIs) using 3D structure information. Compared to existing tools, our implementation leverages GPUs and multi-node parallelism for ultrafast computation. For example, on AIST’s ABCI supercomputer with 512 nodes, we can predict approximately 70 million PPIs per day.
MEGADOCK version 4 is released under a CC BY-NC license for non-commercial use. For commercial applications, please contact us.
GitHub Repository: https://github.com/akiyamalab/MEGADOCK
Try MEGADOCK on Google Colab: https://github.com/ohuelab/MEGADOCK-on-Colab
MEGADOCK is also listed in the NVIDIA GPU Application Catalog.

Example applications of MEGADOCK in PPI research:
- Kami D, et al., PLoS ONE, 2018: Journal website | PubMed
- Hayashi T, et al., BMC Bioinformatics, 2018: Journal website | PubMed | Slide
- Ohue M, et al., Jikkenigaku, 2019: Book website
Peptide Drug Discovery Support
We are developing computational techniques to analyze peptide properties and support drug development, especially for peptide therapeutics, which are attracting attention as the third class of pharmaceuticals.
One example is the development of a method to predict the cell membrane permeability of cyclic peptides. This approach, based on enhanced sampling molecular dynamics simulations, has been validated using ~150 peptides. It addresses a bottleneck in peptide drug design and is expected to contribute significantly to the field. (Supported by the MEXT Regional Innovation Ecosystem Program)
Related Publications:
- Sugita M, et al., J. Chem. Inf. Model., 2022: Journal website | PubMed
- Sugita M, et al., J. Chem. Inf. Model., 2021: Journal website | PubMed
We have newly reported the structure of a cyclic peptide complexed with human serum albumin, revealing the mechanism of its stability in the body. (Joint research with Nagaoka College, Rigaku Corporation, and the University of Tokyo)
- Ito T, et al., J. Med. Chem., 2020: Journal website | bioRxiv | PubMed
Competitive Research Funding
- KAKENHI, Grant-in-Aid for Transformative Research Areas
- KAKENHI, Grant-in-Aid for Scientific Research(B)
- JST FOREST
- AMED BINDS
Collaborative Research with Industry
(Publicly available or permitted for disclosure)
- Alivexis Inc. (formerly Modulus Inc.)
- Development of a novel FEP method for structurally diverse compounds: Press Release
- Research agreement with Tokyo Tech: PR Times
- PeptiDream Inc.
- Joint research on in silico technologies for special peptide drug discovery: Tokyo Tech News
- FUJIFILM Corporation
- Presentation of collaborative research with Ohue Lab at IEEE BIBM 2024 in Lisbon: Twitter Announcement
- Perseus Proteomics Inc.
